pacman::p_load(
rio, # File import
here, # File locator
skimr, # get overview of data
tidyverse, # data management + ggplot2 graphics,
gtsummary, # summary statistics and tests
rstatix, # statistics
corrr, # correlation analayis for numeric variables
janitor, # adding totals and percents to tables
flextable # converting tables to HTML
)18 Simple statistical tests
This page demonstrates how to conduct simple statistical tests using base R, rstatix, and gtsummary.
- T-test
- Shapiro-Wilk test
- Wilcoxon rank sum test
- Kruskal-Wallis test
- Chi-squared test
- Correlations between numeric variables
…many other tests can be performed, but we showcase just these common ones and link to further documentation.
Each of the above packages bring certain advantages and disadvantages:
- Use base R functions to print a statistical outputs to the R Console
- Use rstatix functions to return results in a data frame, or if you want tests to run by group
- Use gtsummary if you want to quickly print publication-ready tables
18.1 Preparation
Load packages
This code chunk shows the loading of packages required for the analyses. In this handbook we emphasize p_load() from pacman, which installs the package if necessary and loads it for use. You can also load installed packages with library() from base R. See the page on R basics for more information on R packages.
Import data
We import the dataset of cases from a simulated Ebola epidemic. If you want to follow along, click to download the “clean” linelist (as .rds file). Import your data with the import() function from the rio package (it accepts many file types like .xlsx, .rds, .csv - see the Import and export page for details).
# import the linelist
linelist <- import("linelist_cleaned.rds")The first 50 rows of the linelist are displayed below.
18.2 base R
You can use base R functions to conduct statistical tests. The commands are relatively simple and results will print to the R Console for simple viewing. However, the outputs are usually lists and so are harder to manipulate if you want to use the results in subsequent operations.
T-tests
A t-test, also called “Student’s t-Test”, is typically used to determine if there is a significant difference between the means of some numeric variable between two groups. Here we’ll show the syntax to do this test depending on whether the columns are in the same data frame.
Syntax 1: This is the syntax when your numeric and categorical columns are in the same data frame. Provide the numeric column on the left side of the equation and the categorical column on the right side. Specify the dataset to data =. Optionally, set paired = TRUE, and conf.level = (0.95 default), and alternative = (either “two.sided”, “less”, or “greater”). Enter ?t.test for more details.
## compare mean age by outcome group with a t-test
t.test(age_years ~ gender, data = linelist)
Welch Two Sample t-test
data: age_years by gender
t = -21.344, df = 4902.3, p-value < 2.2e-16
alternative hypothesis: true difference in means between group f and group m is not equal to 0
95 percent confidence interval:
-7.571920 -6.297975
sample estimates:
mean in group f mean in group m
12.60207 19.53701 Syntax 2: You can compare two separate numeric vectors using this alternative syntax. For example, if the two columns are in different data sets.
t.test(df1$age_years, df2$age_years)You can also use a t-test to determine whether a sample mean is significantly different from some specific value. Here we conduct a one-sample t-test with the known/hypothesized population mean as mu =:
t.test(linelist$age_years, mu = 45)Shapiro-Wilk test
The Shapiro-Wilk test can be used to determine whether a sample came from a normally-distributed population (an assumption of many other tests and analysis, such as the t-test). However, this can only be used on a sample between 3 and 5000 observations. For larger samples a quantile-quantile plot may be helpful.
shapiro.test(linelist$age_years)Wilcoxon rank sum test
The Wilcoxon rank sum test, also called the Mann–Whitney U test, is often used to help determine if two numeric samples are from the same distribution when their populations are not normally distributed or have unequal variance.
## compare age distribution by outcome group with a wilcox test
wilcox.test(age_years ~ outcome, data = linelist)
Wilcoxon rank sum test with continuity correction
data: age_years by outcome
W = 2501868, p-value = 0.8308
alternative hypothesis: true location shift is not equal to 0Kruskal-Wallis test
The Kruskal-Wallis test is an extension of the Wilcoxon rank sum test that can be used to test for differences in the distribution of more than two samples. When only two samples are used it gives identical results to the Wilcoxon rank sum test.
## compare age distribution by outcome group with a kruskal-wallis test
kruskal.test(age_years ~ outcome, linelist)
Kruskal-Wallis rank sum test
data: age_years by outcome
Kruskal-Wallis chi-squared = 0.045675, df = 1, p-value = 0.8308Chi-squared test
Pearson’s Chi-squared test is used in testing for significant differences between categorical croups.
## compare the proportions in each group with a chi-squared test
chisq.test(linelist$gender, linelist$outcome)
Pearson's Chi-squared test with Yates' continuity correction
data: linelist$gender and linelist$outcome
X-squared = 0.0011841, df = 1, p-value = 0.972518.3 rstatix package
The rstatix package offers the ability to run statistical tests and retrieve results in a “pipe-friendly” framework. The results are automatically in a data frame so that you can perform subsequent operations on the results. It is also easy to group the data being passed into the functions, so that the statistics are run for each group.
Summary statistics
The function get_summary_stats() is a quick way to return summary statistics. Simply pipe your dataset to this function and provide the columns to analyse. If no columns are specified, the statistics are calculated for all columns.
By default, a full range of summary statistics are returned: n, max, min, median, 25%ile, 75%ile, IQR, median absolute deviation (mad), mean, standard deviation, standard error, and a confidence interval of the mean.
linelist %>%
rstatix::get_summary_stats(age, temp)# A tibble: 2 × 13
variable n min max median q1 q3 iqr mad mean sd se
<fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 age 5802 0 84 13 6 23 17 11.9 16.1 12.6 0.166
2 temp 5739 35.2 40.8 38.8 38.2 39.2 1 0.741 38.6 0.977 0.013
# ℹ 1 more variable: ci <dbl>You can specify a subset of summary statistics to return by providing one of the following values to type =: “full”, “common”, “robust”, “five_number”, “mean_sd”, “mean_se”, “mean_ci”, “median_iqr”, “median_mad”, “quantile”, “mean”, “median”, “min”, “max”.
It can be used with grouped data as well, such that a row is returned for each grouping-variable:
linelist %>%
group_by(hospital) %>%
rstatix::get_summary_stats(age, temp, type = "common")# A tibble: 12 × 11
hospital variable n min max median iqr mean sd se ci
<chr> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Central Hos… age 445 0 58 12 15 15.7 12.5 0.591 1.16
2 Central Hos… temp 450 35.2 40.4 38.8 1 38.5 0.964 0.045 0.089
3 Military Ho… age 884 0 72 14 18 16.1 12.4 0.417 0.818
4 Military Ho… temp 873 35.3 40.5 38.8 1 38.6 0.952 0.032 0.063
5 Missing age 1441 0 76 13 17 16.0 12.9 0.339 0.665
6 Missing temp 1431 35.8 40.6 38.9 1 38.6 0.97 0.026 0.05
7 Other age 873 0 69 13 17 16.0 12.5 0.422 0.828
8 Other temp 862 35.7 40.8 38.8 1.1 38.5 1.01 0.034 0.067
9 Port Hospit… age 1739 0 68 14 18 16.3 12.7 0.305 0.598
10 Port Hospit… temp 1713 35.5 40.6 38.8 1.1 38.6 0.981 0.024 0.046
11 St. Mark's … age 420 0 84 12 15 15.7 12.4 0.606 1.19
12 St. Mark's … temp 410 35.9 40.6 38.8 1.1 38.5 0.983 0.049 0.095You can also use rstatix to conduct statistical tests:
T-test
Use a formula syntax to specify the numeric and categorical columns:
linelist %>%
t_test(age_years ~ gender)# A tibble: 1 × 10
.y. group1 group2 n1 n2 statistic df p p.adj p.adj.signif
* <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <dbl> <chr>
1 age_… f m 2807 2803 -21.3 4902. 9.89e-97 9.89e-97 **** Or use ~ 1 and specify mu = for a one-sample T-test. This can also be done by group.
linelist %>%
t_test(age_years ~ 1, mu = 30)# A tibble: 1 × 7
.y. group1 group2 n statistic df p
* <chr> <chr> <chr> <int> <dbl> <dbl> <dbl>
1 age_years 1 null model 5802 -84.2 5801 0If applicable, the statistical tests can be done by group, as shown below:
linelist %>%
group_by(gender) %>%
t_test(age_years ~ 1, mu = 18)# A tibble: 3 × 8
gender .y. group1 group2 n statistic df p
* <chr> <chr> <chr> <chr> <int> <dbl> <dbl> <dbl>
1 f age_years 1 null model 2807 -29.8 2806 7.52e-170
2 m age_years 1 null model 2803 5.70 2802 1.34e- 8
3 <NA> age_years 1 null model 192 -3.80 191 1.96e- 4Shapiro-Wilk test
As stated above, sample size must be between 3 and 5000.
linelist %>%
head(500) %>% # first 500 rows of case linelist, for example only
shapiro_test(age_years)# A tibble: 1 × 3
variable statistic p
<chr> <dbl> <dbl>
1 age_years 0.917 6.67e-16Wilcoxon rank sum test
linelist %>%
wilcox_test(age_years ~ gender)# A tibble: 1 × 9
.y. group1 group2 n1 n2 statistic p p.adj p.adj.signif
* <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <chr>
1 age_years f m 2807 2803 2829274 3.47e-74 3.47e-74 **** Kruskal-Wallis test
Also known as the Mann-Whitney U test.
linelist %>%
kruskal_test(age_years ~ outcome)# A tibble: 1 × 6
.y. n statistic df p method
* <chr> <int> <dbl> <int> <dbl> <chr>
1 age_years 5888 0.0457 1 0.831 Kruskal-WallisChi-squared test
The chi-square test function accepts a table, so first we create a cross-tabulation. There are many ways to create a cross-tabulation (see Descriptive tables) but here we use tabyl() from janitor and remove the left-most column of value labels before passing to chisq_test().
linelist %>%
tabyl(gender, outcome) %>%
select(-1) %>%
chisq_test()# A tibble: 1 × 6
n statistic p df method p.signif
* <dbl> <dbl> <dbl> <int> <chr> <chr>
1 5888 3.53 0.473 4 Chi-square test ns Many many more functions and statistical tests can be run with rstatix functions. See the documentation for rstatix online here or by entering ?rstatix.
18.4 gtsummary package
Use gtsummary if you are looking to add the results of a statistical test to a pretty table that was created with this package (as described in the gtsummary section of the Descriptive tables page).
Performing statistical tests of comparison with tbl_summary is done by adding the add_p function to a table and specifying which test to use. It is possible to get p-values corrected for multiple testing by using the add_q function. Run ?tbl_summary for details.
Chi-squared test
Compare the proportions of a categorical variable in two groups. The default statistical test for add_p() when applied to a categorical variable is to perform a chi-squared test of independence with continuity correction, but if any expected call count is below 5 then a Fisher’s exact test is used.
linelist %>%
select(gender, outcome) %>% # keep variables of interest
tbl_summary(by = outcome) %>% # produce summary table and specify grouping variable
add_p() # specify what test to perform1323 observations missing `outcome` have been removed. To include these observations, use `forcats::fct_na_value_to_level()` on `outcome` column before passing to `tbl_summary()`.Characteristic |
Death, N = 2,582 1 |
Recover, N = 1,983 1 |
p-value 2 |
|---|---|---|---|
| gender | >0.9 | ||
| f | 1,227 (50%) | 953 (50%) | |
| m | 1,228 (50%) | 950 (50%) | |
| Unknown | 127 | 80 | |
|
1
n (%) |
|||
|
2
Pearson’s Chi-squared test |
|||
T-tests
Compare the difference in means for a continuous variable in two groups. For example, compare the mean age by patient outcome.
linelist %>%
select(age_years, outcome) %>% # keep variables of interest
tbl_summary( # produce summary table
statistic = age_years ~ "{mean} ({sd})", # specify what statistics to show
by = outcome) %>% # specify the grouping variable
add_p(age_years ~ "t.test") # specify what tests to perform1323 observations missing `outcome` have been removed. To include these observations, use `forcats::fct_na_value_to_level()` on `outcome` column before passing to `tbl_summary()`.Characteristic |
Death, N = 2,582 1 |
Recover, N = 1,983 1 |
p-value 2 |
|---|---|---|---|
| age_years | 16 (12) | 16 (13) | 0.6 |
| Unknown | 32 | 28 | |
|
1
Mean (SD) |
|||
|
2
Welch Two Sample t-test |
|||
Wilcoxon rank sum test
Compare the distribution of a continuous variable in two groups. The default is to use the Wilcoxon rank sum test and the median (IQR) when comparing two groups. However for non-normally distributed data or comparing multiple groups, the Kruskal-wallis test is more appropriate.
linelist %>%
select(age_years, outcome) %>% # keep variables of interest
tbl_summary( # produce summary table
statistic = age_years ~ "{median} ({p25}, {p75})", # specify what statistic to show (this is default so could remove)
by = outcome) %>% # specify the grouping variable
add_p(age_years ~ "wilcox.test") # specify what test to perform (default so could leave brackets empty)1323 observations missing `outcome` have been removed. To include these observations, use `forcats::fct_na_value_to_level()` on `outcome` column before passing to `tbl_summary()`.Characteristic |
Death, N = 2,582 1 |
Recover, N = 1,983 1 |
p-value 2 |
|---|---|---|---|
| age_years | 13 (6, 23) | 13 (6, 23) | 0.8 |
| Unknown | 32 | 28 | |
|
1
Median (IQR) |
|||
|
2
Wilcoxon rank sum test |
|||
Kruskal-wallis test
Compare the distribution of a continuous variable in two or more groups, regardless of whether the data is normally distributed.
linelist %>%
select(age_years, outcome) %>% # keep variables of interest
tbl_summary( # produce summary table
statistic = age_years ~ "{median} ({p25}, {p75})", # specify what statistic to show (default, so could remove)
by = outcome) %>% # specify the grouping variable
add_p(age_years ~ "kruskal.test") # specify what test to perform1323 observations missing `outcome` have been removed. To include these observations, use `forcats::fct_na_value_to_level()` on `outcome` column before passing to `tbl_summary()`.Characteristic |
Death, N = 2,582 1 |
Recover, N = 1,983 1 |
p-value 2 |
|---|---|---|---|
| age_years | 13 (6, 23) | 13 (6, 23) | 0.8 |
| Unknown | 32 | 28 | |
|
1
Median (IQR) |
|||
|
2
Kruskal-Wallis rank sum test |
|||
18.5 Correlations
Correlation between numeric variables can be investigated using the tidyverse
corrr package. It allows you to compute correlations using Pearson, Kendall tau or Spearman rho. The package creates a table and also has a function to automatically plot the values.
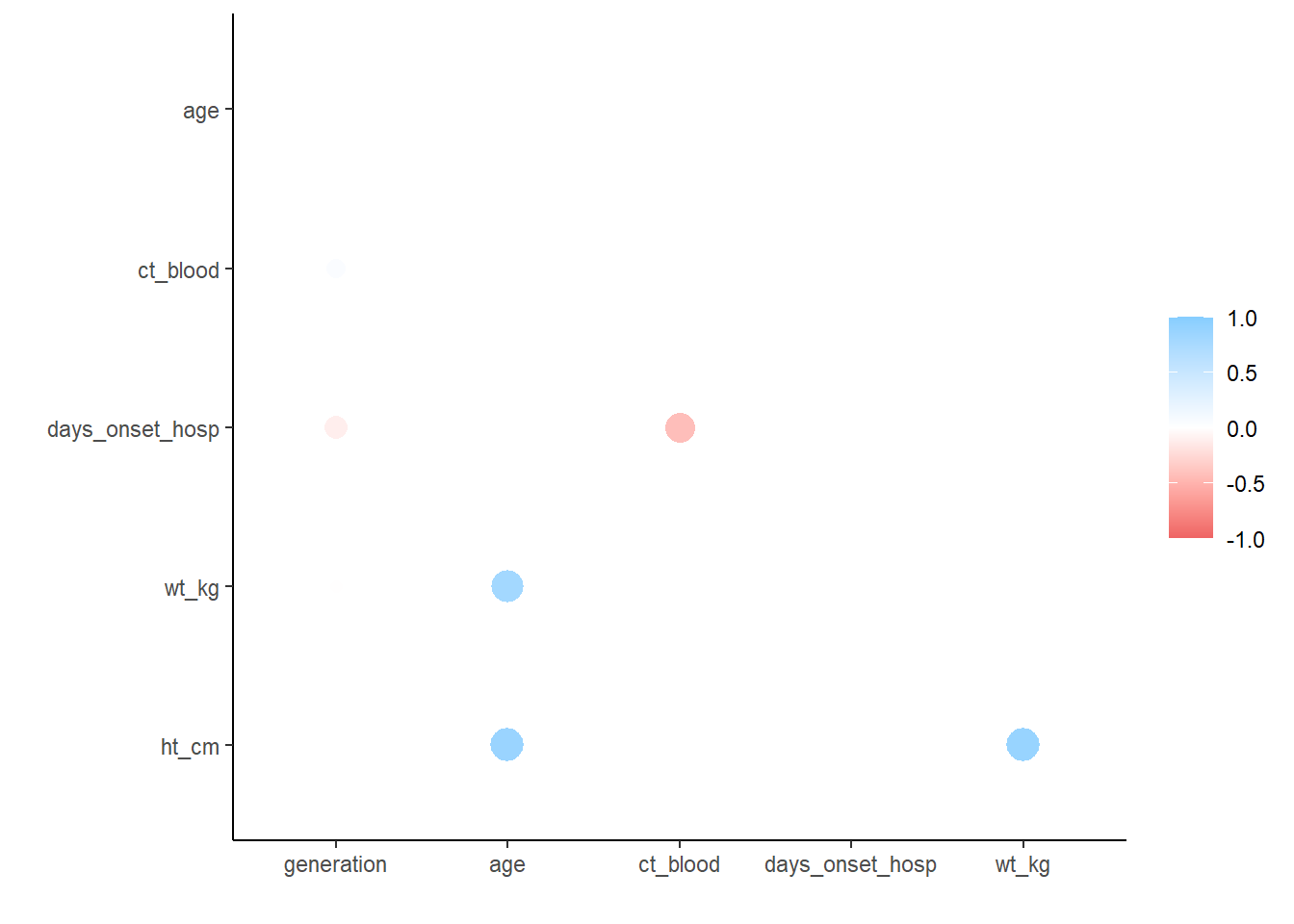
correlation_tab <- linelist %>%
select(generation, age, ct_blood, days_onset_hosp, wt_kg, ht_cm) %>% # keep numeric variables of interest
correlate() # create correlation table (using default pearson)
correlation_tab # print# A tibble: 6 × 7
term generation age ct_blood days_onset_hosp wt_kg ht_cm
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 generation NA -2.22e-2 0.179 -0.288 -0.0302 -0.00942
2 age -0.0222 NA 0.00849 -0.000635 0.833 0.877
3 ct_blood 0.179 8.49e-3 NA -0.600 -0.00636 0.0181
4 days_onset_hosp -0.288 -6.35e-4 -0.600 NA 0.0153 -0.00953
5 wt_kg -0.0302 8.33e-1 -0.00636 0.0153 NA 0.884
6 ht_cm -0.00942 8.77e-1 0.0181 -0.00953 0.884 NA ## remove duplicate entries (the table above is mirrored)
correlation_tab <- correlation_tab %>%
shave()
## view correlation table
correlation_tab# A tibble: 6 × 7
term generation age ct_blood days_onset_hosp wt_kg ht_cm
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 generation NA NA NA NA NA NA
2 age -0.0222 NA NA NA NA NA
3 ct_blood 0.179 0.00849 NA NA NA NA
4 days_onset_hosp -0.288 -0.000635 -0.600 NA NA NA
5 wt_kg -0.0302 0.833 -0.00636 0.0153 NA NA
6 ht_cm -0.00942 0.877 0.0181 -0.00953 0.884 NA## plot correlations
rplot(correlation_tab)18.6 Resources
Much of the information in this page is adapted from these resources and vignettes online: